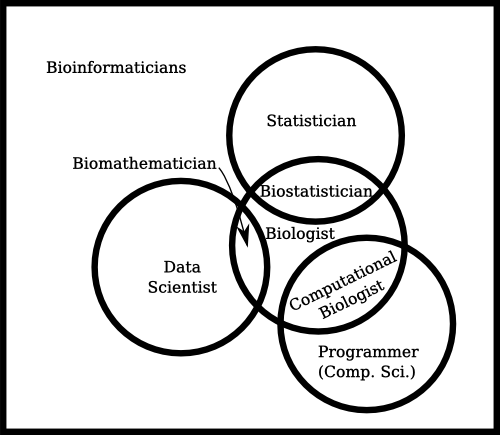
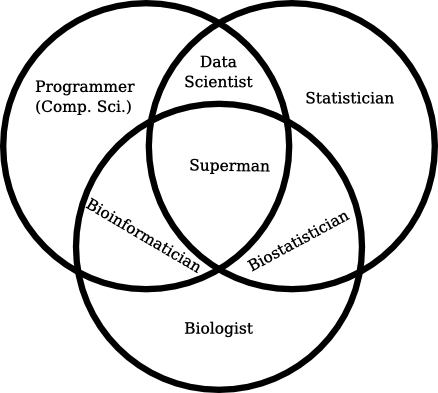
I’ve been following the bioinformatics sub-reddit for the past couple of months, ever since I stumbled upon it when a colleague asked me about bioinformatics resources on the web. It’s a fascinating place to visit, but it’s incredibly repetitive in that people keep asking “How do I become a bioinformatician?”
Unfortunately there is not a single answer, because bioinformatics isn’t a single job – it’s a collection of people who have found a way to live with one foot in each of two worlds: computer programming and biology. Getting a firm footing in each can be a serious challenge, as people spend years studying just one of those to become proficient at it.
However, I think there are some common threads that tie the field together. You need to invest the time in at least a handful of basic fields: some basic programming, some elementary cell biology and at least a simple understanding of math or statistics. What you can accomplish with just that little can be incredibly productive. Mostly in terms of automation of data processing or modelling of your results.
On the other hand, bioinformatics also includes a lot of sub-disciplines. Great programmers can build incredible pipelines. Great mathematicians can invent or apply algorithms to create new ways of interpreting data, and great biologists can develop heuristics and re-interpret data in new ways to generate insights that others have overlooked. There’s even room for “neat freaks” in organizing and imposing order on unruly data.
The challenge of becoming a bioinformatician is learning where your strengths and weaknesses lay, and using them to your advantage. Finding a research group that shores up your weaknesses – or helps you fill them in – can be a great boost to your career. After my masters degree, I felt I had two big gaping holes in my resume: big data and databases, which I made the focus of my PhD research. Coming out of my defence, I felt I was able to bring a more balanced approach to the table – and had simultaneously purged any instinct I might have ever had to reach for a spreadsheet to interpret information. (Spreadsheets and big data don’t mix.)
So, where does that lead an aspiring bioinformatician? Unless you take the time to do both a computer science degree and a biology degree, you probably won’t be able to shoehorn everything in to become an expert in both, and not everyone wants to get their PhD to fill in the gaps left in an undergrad education.
With that said, let me lay down a few useful points:
- Pick and chose to study subjects that interest you because you’ll at least end up with strengths in things you enjoy, which leads to jobs doing things you enjoy.
- You can always learn something new later… but take opportunities to try new things when they come.
- Remember that you’re not going to be the expert in every field you put your foot into – so look for opportunities to collaborate with the people who are. (If you’re going into bioinformatics and expect to do everything yourself, you’re probably doing it wrong.)
- Don’t be afraid of the fact that you don’t know stuff. Your job isn’t to be the best biologist and best computer scientist at the same time – it’s to be the bridge between. The stronger your foundations, the better a bridge you can be, but unlike a concrete bridge, you can always invest in learning more.
- Yes, higher education does help in this field. Bioinformatics is still dominated by research based organizations, and the academic hierarchy saturates the mindset of bioinformaticians everywhere. (Or, almost everywhere.)
- Bioinformatics is also about the “soft” skills. Don’t forget that bioinformaticians are also in a good place to be good leaders – since you’ll be one of the few people who can speak both languages, and tie together groups that would otherwise lack a common language.
- Don’t believe the hype about what you should learn: R isn’t really the only language for doing bioinformatics. Perl isn’t always evil (just most of the time, though it did save the human genome…), Java isn’t the slowest language out there, and c isn’t only for hardcore programmers. (Python, though, is a pretty good all-around language.) Everyone has an opinion on where bioinformatics is going – but it’s just an opinion, so make your own choices.
At the end of the day, I always give students the same piece of advice: As you go through life, you will learn new skills that you can apply as you see fit. At the end of the day, each of these skills will be a tool in your toolbox that you can turn to when you hit a problem. If you only have a hammer in your toolbox, your repertoire is pretty limited. On the other hand, if you collect a fantastic assembly of tools, you’ll be equipped to handle just about anything that comes your way. Your job is to invest your time into building the best toolkit you can, so that when you get out of school, you’ll be ready to solve as many problems as you can.
Bioinformatics is just a special case of toolbox building, in that you need the tools of at least two disciplines in your toolbox. What you chose to put into your toolbox is entirely up to you, but (to stretch the toolbox analogy just a little too far), take a few minutes to ask if you’d like to be a plumber or a carpenter before you start collecting your tools. Or, without the metaphoric toolkit, ask yourself what kind of bioinformatician you want to be.
Once you know the answer to that question, you’ll figure out pretty quickly which tools you want to start collecting. And the path towards becoming a bioinformatician will start to become clear. It may not take you where you expect, but I can guarantee that you’ll be walking down an interesting road.