I’ve been participating in an interesting conversation on linkedin, which has re-opened the age old question of what is a bioinformatician, which was inspired by a conversation on twitter, that was later blogged. Hopefully I’ve gotten that chain down correctly.
In any case, it appears that there are two competing schools of thought. One is that bioinformatician is a distinct entity, and the other is that it’s a vague term that embraces anyone and anything that has to do with either biology or computer science. Frankly, I feel the second definition is a waste of a perfectly good word, despite being a commonly accepted method.
That leads me to the following two illustrations.
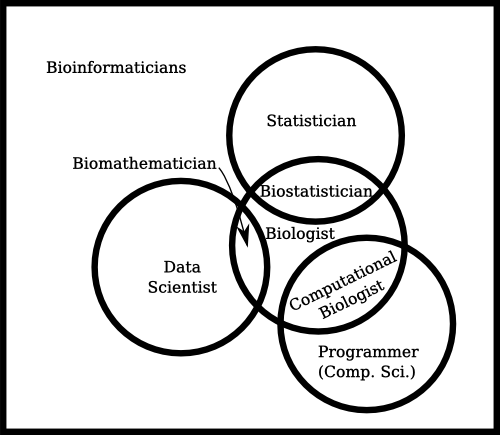
How bioinformatics is often used, and I would argue that it’s being used incorrectly.:
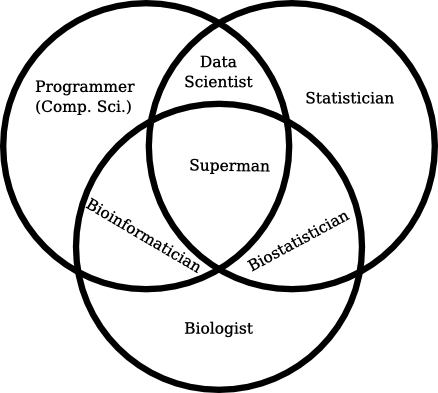
And how it should be used, according to me:
I think the second clearly describes something that just isn’t captured otherwise. It covers a specific skill set that’s otherwise not captured by anything else.
In fact, I have often argued that bioinformatician is really a position along a gradient from computer science to biology, where your skills in computer science would determine whether you’re a computational biologist (someone who applies computer programs to solve biology problems) or a bioinformatician (someone who designs computer programs to solve biology problems). Those, to me, are entirely different skill sets – and although bioinformaticians are often those who end up implementing the computer programs, that’s yet another skill, but can be done by a programmer who doesn’t understand the biology.
That, effectively, makes bioinformatician an accurate description of a useful skill set – and further divides the murky field of “people who understand biology and use computers” – which is vague enough to include people who use an excel spreadsheets to curate bacterial strain collections.
I suppose the next step is to get those who do taxonomy into the computational side of things and have them sort us all out.